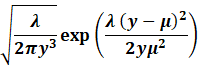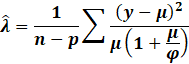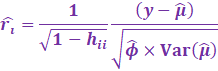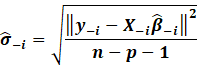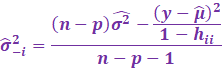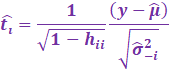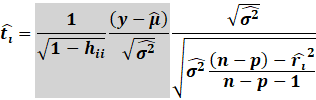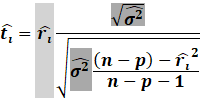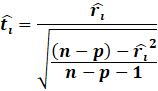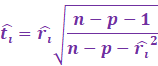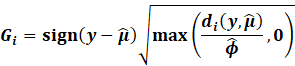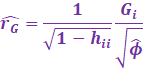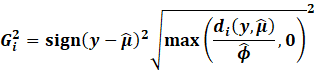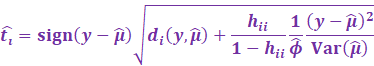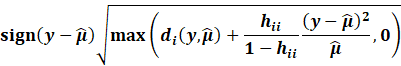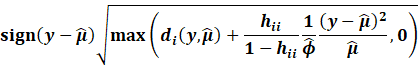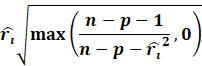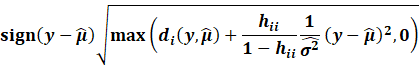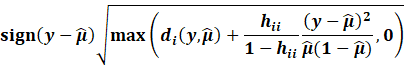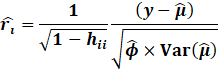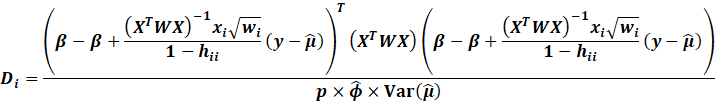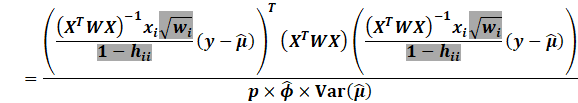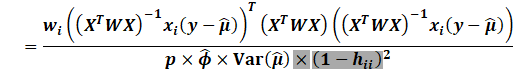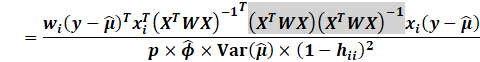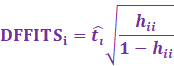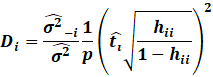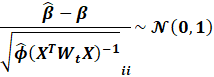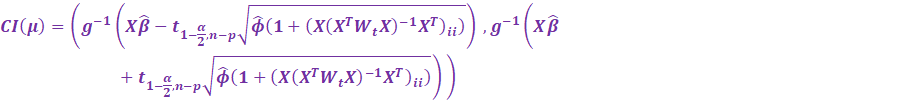Diagnostics for GLMs
How does one find outliers, influential
observations?
How about prediction intervals, confidence
intervals?
https://www.sagepub.com/sites/default/files/upm-binaries/21121_Chapter_15.pdf
[Textbook]
Leverage Score
Estimation
https://icml.cc/2012/papers/80.pdf [2012
Fast approximation of matrix coherence and statistical leverage]
Firstly, we have to derive the hat matrix
for GLMs. We find that:
|
|
Also notice the sum of the leverage scores is equal to the number
of parameters in the model.
[BE CAREFUL NOT ALWAYS!!! Ie like in
Ridge Regression]
However
be careful! The TRUE hat matrix is NOT the formula above, but rather:
|
|
Ive
confirmed the norm between them is off by around p - rank(H).
However, since we only want the leverage scores, we use the trace
property cycling property.
|
|
However, on further notice, we notice via the Cholesky
Decomposition that:
|
|
All in all, this optimized routine computes both the
variances(beta) and leverage(X) optimally in around:
np^2 + 1/3p^3 + 1/2p^3 + p^2 + np^2 + np + n FLOPS.
Or around:
(2/3)p^3 + (2n)p^2 + (n+1)p FLOPS.
And peak memory consumption:
n/Bp + p^2 + n + p SPACE.
Where B is the batch size.
Now going about estimating the leverage scores or the diagonal hat
matrix is a pain. One clearly will be crazy on computing
the full hat matrix then only extracting the diagonals.
Instead sketching comes to the rescue again!
|
|
Also notice the maximum leverage score must be 1 [leverage scores
are between 0 and 1.]
Also, notice the scaling of the normal distribution sketch.
Now the sketching matrices can be the count sketch matrix.
S2 should NOT be a count sketch matrix, since during the
triangular solve.
Notice the error rate. That means in general if the sketching size
requires too many observations,
you would rather use the original matrix.
Standardized,
Studentized Residuals
Pearson residuals are
the per observation component of the Pearson goodness of fit test ie:
|
|
Where we have for the exponential family:
|
Name |
Distribution |
Range |
|
|
|
Poisson |
|
|
|
|
|
Quasi Poisson |
|
|
|
|
|
Gamma |
|
|
|
|
|
Bernoulli |
|
|
|
|
|
Inverse Gaussian |
|
|
|
|
|
Negative
Binomial |
|
|
|
|
Standardized Pearson Residuals
then correct for the leverage scores:
[These are also called internally studentized residuals]
|
|
For practical models:
|
Name |
Range |
|
|
|
|
Poisson |
|
|
|
|
|
Quasi Poisson |
|
|
|
|
|
Linear |
|
|
|
|
|
Log Linear |
|
|
|
|
|
ReLU Linear |
|
|
|
|
|
CLogLog Logistic |
|
|
|
|
|
Logistic |
|
|
|
|
The PRESS residuals showcase the difference in the
prediction and true value if we
excluded the observation from the model.
These can be proved by using the Sherman Morrison Inverse
Identity formula.
|
|
However, in outlier analysis, one generally considers the externally
studentized residuals,
which essentially is when the variance is estimated whenever each
observation is deleted it.
Minus 1 because one datapoint is deleted each time.
|
|
Now to simplify this, we utilize the PRESS residuals:
[ Gray part skips steps which are a bit tedious to showcase 😊 ]
|
|
Then the externally studentized residuals utilises this new
excluded variance:
|
|
However in GLMs, we cannot resort to this estimate for the OLS
model.
Rather, we first remember the Deviance Residuals:
|
|
The standardized deviance residuals are then:
|
|
Then, we get the Williams approximation to externally
studentized residuals:
|
|
Which we find that:
|
|
For practical models:
|
Name |
|
|
|
|
|
Poisson |
|
|
|
|
|
Quasi Poisson |
|
|
|
|
|
Linear |
|
|
|
|
|
Log Linear |
|
|
|
|
|
ReLU Linear |
|
|
|
|
|
|
|
|
|
|
|
Logistic |
|
|
|
|
Cooks Distance,
DFFITS
Popular methods to detect influential or possible outliers include
the Cooks Distance and DFFITS.
|
|
The Cooks Distance is essentially the total change in MSE if an
observation is removed.
It shows how much error can be seen in standard deviation units on
how the error changes
if an observation is removed.
|
|
First notice by using the Sherman Morrison inverse formula that:
|
|
We then expand:
|
|
Generally, a cutoff of 4/n is reasonable. Likewise, one can use
the 50% percentile of the F distribution.
Though for large n, this will converge to 1.
On the other hand, DFFITS is a similar measure to the Cooks
Distance.
DFFITS rather uses the externally studentized residual, which is
more preferred than the Cooks Distance.
|
|
In fact, DFFITS is closely related to the Cooks Distance:
|
|
The cutoff value for DFFITS is:
|
|
Finally to plot and showcase the possible outliers, one can use
for both axes:
|
|
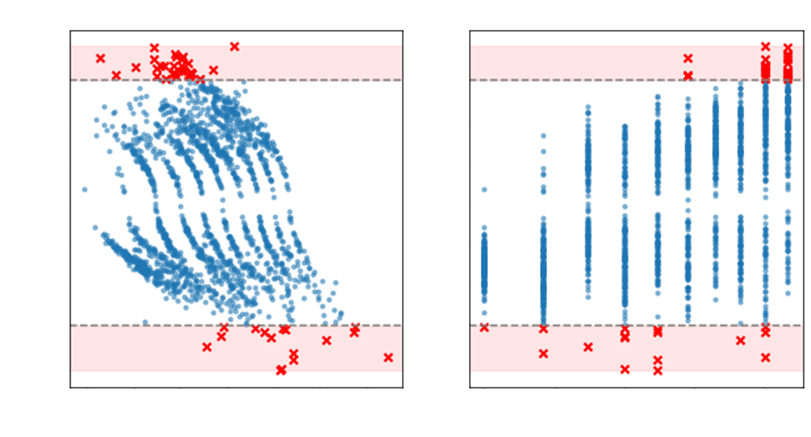
However in practice, the identification of outliers is more
complicated than using DFFITS, since DFFITS can suggest
too many outlier candidates. Rather we use the Studentized
Residuals vs Leverage plot:
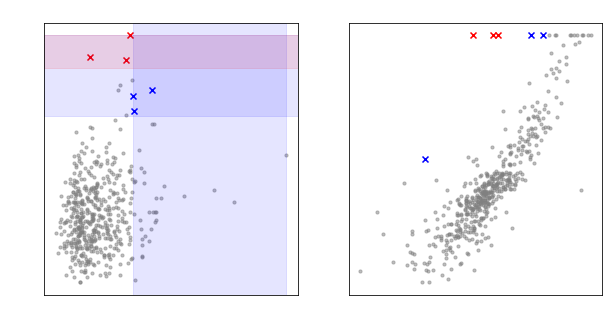
The cutoff is 3 * the studentized residual (ie the residual is 3
standard deviations away from the mean).
Likewise the leverage much be at least 2 times the average
leverage.
|
|
However, we need to also capture wildly off-putting results. So, we
also include anything above 5 standard deviations.
|
|
Confidence,
Prediction Intervals
Confidence and Prediction Intervals for both new data and old data
are super important aspects.
The trick is one utilises the leverage scores.
Remember that:
|
|
This essentially means that:
|
|
Notice how this is NOT the leverage
score!!!!!!
And then for GLMs, via the inverse link function (the activation
function):
|
|
Then, we have the upper
(97.5%) and lower (2.5%) confidence
intervals:
|
|
Be careful! Its the upper
(97.5%) and lower (2.5%) confidence
intervals! Not 95%.
For new data points though, we have to estimate the factor. We can
do this via:
[Where A is the new data matrix, psi is the new predicted value
and z is the new weight]
|
|
Remember though previously we performed the Cholesky Decomposition
on the variance matrix.
So, we can speed things up by noticing:
[Notice we exclude the new weights]
|
|
For Prediction Intervals, we shall not prove why, but there is in
fact a +1 factor inside the leverage square root:
|
|
Notice if the dispersion parameter is 1 (Poisson, Bernoulli
models), then the normal distribution is
used instead of the Student T distribution for the critical
values.
Copyright Daniel Han 2024. Check out Unsloth!