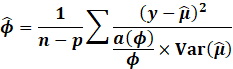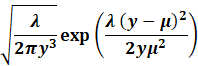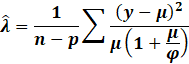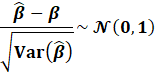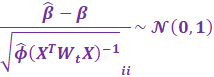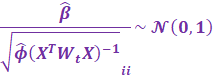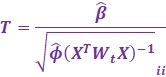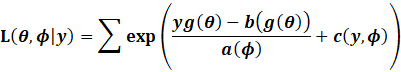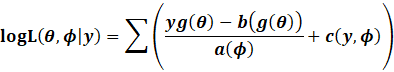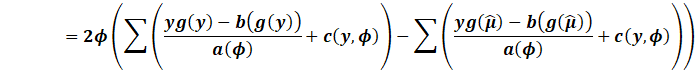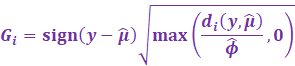Statistical Inference
for GLMs
After carefully fitting the coefficients for a GLM,
one then must test the significance of them.
We can easily test them via statistical tests.
https://www.sagepub.com/sites/default/files/upm-binaries/21121_Chapter_15.pdf
[Textbook]
Dispersion Parameter
Estimation
Recall that:
|
|
This means that the final variance of the parameters is:
[Notice W is not weights, but rather the original variance, since
we do not cancel the dispersion out]
|
|
However, we do not know the dispersion parameters. So, we estimate
them via the method of moments:
|
|
Now lets derive the variance for the
generic exponential families:
|
Distribution |
Range |
|
|
|
|
Poisson |
|
|
|
|
|
Gaussian |
|
|
|
|
|
Gamma |
|
|
|
|
|
Bernoulli |
|
|
|
|
|
Inverse Gaussian |
|
|
|
|
|
Negative Binomial |
|
|
|
|
Notice for the Poisson model, to counteract over or under
dispersion, we generally let the dispersion parameter to be:
|
|
This is called the Quasi Poisson Model
Coefficient
Significance
A Wald Test can be used to determine the significance of
coefficients, assuming CLT:
|
|
To test the hypothesis that the coefficients are zero,
|
|
Given a 0.05% significance level, we then perform a two sided Z test:
|
s = 2 * (1 - scipy.stats.norm.cdf(abs(T))) |
However, for when a dispersion parameter is estimated, one must
use a 2 sided T test.
|
s = 2 * (1 - scipy.stats.t.cdf(abs(T),
df = n - p)) |
Likewise for
confidence intervals at the 0.05 significance level, we show both Z and T
tests.
|
k = scipy.stats.norm.ppf(1
- 0.05/2)
k = scipy.stats.t.ppf(1
- 0.05/2, df = n - p) |
Then, we have the upper
(97.5%) and lower (2.5%) confidence
intervals:
|
|
Be careful! It’s the upper (97.5%) and lower (2.5%) confidence
intervals! Not 95%.
In general, if the sample size is large enough, the T distribution
converges to a Normal distribution.
At the 0.05 significance level, we have that
|
|
However, with the T distribution on less than 10,000 samples, the
value is much larger ie:
|
|
So, one should keep a table of precomputed values less than 10,000
or so samples,
then clip the value to 1.96.
So finally, we have the following
|
Name |
Distribution |
Range |
|
|
Test Statistic |
|
Poisson |
|
|
|
|
|
|
Quasi Poisson |
|
|
|
|
|
|
Gaussian |
|
|
|
|
|
|
Gamma |
|
|
|
|
|
|
Bernoulli |
|
|
|
|
|
|
Inverse Gaussian |
|
|
|
|
|
|
Negative
Binomial |
|
|
|
|
|
However, for the Quasi Poisson Model, one uses a T
distribution.
Degrees of Freedom
Estimation
In general inference of many parameters, n-p is taken to be the
degree of freedom.
However, in statsmodels and many other
statistical packages, rather the actual rank of the matrix is used.
|
|
However, when Cholesky or even the modified Cholesky
(Levenberg–Marquardt) algorithm is used, one does not
attain the singular values.
Pivoted Cholesky can be used to attain an approximate rank.
Likewise, entries which are negative or zero
in the factorization can be considered as rank deficient columns.
Variance Estimation
If the data matrix is exceptionally large, one cannot afford to
invert the matrix.
So, we have to find an approximate
inverse to the matrix.
The most basic approximation is to just use the diagonal of the weights matrix ie:
|
|
However, the error especially for p-value computations is very very bad.
One can also use the Power Method to approximate the top K
eigenvalues and eigenvectors, say like 5,
then use these and compute the diagonals ie:
[Notice the clipping of the eigenvalues]
|
|
However, the power method find the largest
eigenvalues. Since we want the inverse matrix, we rather
want the SMALLEST eigenvalues.
We can do this via the inverse power method.
|
|
However, given a large matrix, it becomes infeasible to even solve
the system of equations.
One can then use MINRES to solve the system. However, MINRES has
bad convergence qualities.
One should rather use LGMRES, which fixes MINRES and converges
much faster.
However, this is still prohibitive. Instead sketching comes to the
rescue. Assuming weights are positive, we have that:
|
|
So the algorithm becomes:
|
|
Likelihood and
Deviance
The deviance is the measure of relative importance when comparing
2 models.
|
|
The first term is called the saturated model.
This is when we create a new linear model where the predictions
are the actual targets themselves!
Notice for the exponential family:
|
|
So, for the exponential family:
|
|
|
|
|
|
|
|
|
Poisson |
|
|
|
|
|
|
|
Quasi Poisson |
|
|
|
|
|
|
|
Gaussian |
|
|
|
|
|
|
|
Gamma |
|
|
|
|
|
|
|
Bernoulli |
|
|
|
|
|
|
|
Inverse Gaussian |
|
|
|
|
|
|
|
Negative Binomial |
|
|
|
|
|
|
And for practical models:
|
Name |
Range |
|
|
|
|
|
|
Poisson |
|
|
|
|
|
|
|
Quasi Poisson |
|
|
|
|
|
|
|
Linear |
|
|
|
|
|
|
|
Log Linear |
|
|
|
|
|
|
|
ReLU Linear |
|
|
|
|
|
|
|
CLogLog Logistic |
|
|
|
|
|
|
|
Logistic |
|
|
|
|
|
|
The residual deviance is the difference between this
saturated model and the fitted model itself.
|
|
For the simple linear regression model we
have that:
|
|
And for practical models, the case wise deviances:
|
Name |
Range |
|
|
|
|
|
|
|
Poisson |
|
|
|
|
|
|
|
|
Quasi Poisson |
|
|
|
|
|
|
|
|
Linear |
|
|
|
|
|
|
|
|
Log Linear |
|
|
|
|
|
|
|
|
ReLU Linear |
|
|
|
|
|
|
|
|
CLogLog Logistic |
|
|
|
|
|
|
|
|
Logistic |
|
|
|
|
|
|
|
And finally, the deviance residuals are then calculated as:
|
|
And the deviance score is calculated as:
|
|
AIC, BIC
https://en.wikipedia.org/wiki/Akaike_information_criterion
[AIC]
https://en.wikipedia.org/wiki/Bayesian_information_criterion [BIC]
The Akaike Information Criterion determines if a model is a good
fit and is not overfit to the data.
It measures the models strengths, and can
be used as a way to compare other models.
Choose the LOWEST AIC value.
|
|
AIC can be used in the relative likelihood ratio test,
where we can compare models.
Say we have 10 models, with the minimum AIC score as MIN.
Then the quantity
|
|
Can be used to determine the probability another model can
minimise the information criterion.
Say if the minimum was 100, and another model had an AIC of 102.
|
|
This means model 2 has a 36.8% chance of minimising AIC rather
than the best model.
The strength of a model can be evaluated as follows:
|
|
Comparison |
|
|
Hopeless |
|
|
Positive |
|
|
Strong |
|
|
Very strong |
The BIC or Bayesian Information Criterion is more
advocated since it penalises model complexity.
|
|
For extremely large data, high dimensional BIC is rather used,
where:
|
|
Gamma be can any number greater than 0. Generally set it to zero.
The strength of a model can be evaluated as follows:
|
|
Comparison |
|
|
Hopeless |
|
|
Positive |
|
|
Strong |
|
|
Very strong |
Copyright Daniel Han 2024. Check out Unsloth!