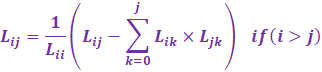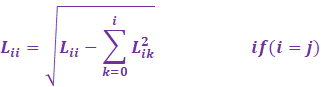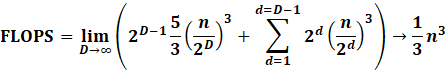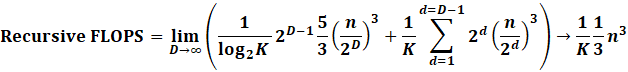Linear
Regression & Least Squares
How does one estimate the parameters of a linear
model?
How about when a matrix is underdetermined or ill
conditioned?
How do we determine the best L2 regularization
parameter?
Just a refresher for overdetermined systems:
|
|
For underdetermined systems:
|
|
Recursive
Stable Cholesky Decomposition
https://en.wikipedia.org/wiki/Cholesky_decomposition [Wikipedia]
https://en.wikipedia.org/wiki/Denormal_number#Performance_issues
[Wikipedia]
https://en.wikipedia.org/wiki/Schur_complement [Wikipedia]
We have already showcased how using Cholesky is
paramount to superior speed.
This is because 2 triangular solves (P^2) can be
used:
|
|
However, actually performing
the Cholesky decomposition has many issues.
The first issue is how does one actually
compute the upper triangular factor U?
From Wikipedia, the Cholesky Crout
algorithm proceeds column wise:
|
|
In code:
|
for (int i = 0; i < n; i++) { for (int j = 0; j < i;
j++) { float s =
0; for (int
k = 0; k < j; k++) s +=
A[i, k] * A[j, k]; A[i, j] = (A[i, j] - s) / A[i, i]; } float s = 0; for (int k =
0; k < i; k++) s += A[i, k] * Ai[i, k]; A[i, i] = sqrt(A[i, i] - s); } |
However, we can optimize the code by noticing that
our matrix was originally Fortran Contiguous, so each column is
accessed optimally. Likewise, store the divisions
and use multiplication instead.
|
for (int i = 0; i < n; i++) { float *__restrict Ai = &A(i, 0); for (int j =
0; j < i; j++) { float *__restrict Aj =
&A(j, 0); float s =
0; for (int
k = 0; k < j; k++) s +=
Ai[k] * Aj[k]; Ai[j] = div[j] * (Ai[j] - s); } float s = 0; for (int k =
0; k < i; k++) s +=
Ai[k] * Ai[k]; const float aii = sqrt(Ai[i] - s); Ai[i]
= aii; div[i]
= 1/aii; } |
The issue is that the above has many issues, namely
the highlighted.
The first issue is that the square root can become
negative. These also indicate rank deficient columns.
We counteract this by adding a check. We also
notice that using FLT_MAX is SLOW due to underflow and denormals.
(Wikipedia says it can be 100 times slower). And
finally, we have to set all rank deficient columns to
zero.
So, we rather use 1/EPS which is around 8.3e+6
compared to FLT_MAX which is 3.4e+38.
Likewise, we set all elements smaller than EPS to
0.
|
for (int i = 0; i < n; i++) { float
*__restrict Ai = &A(i, 0); for (int j =
0; j < i; j++) { float *__restrict Aj
= &A(j, 0); float s =
0; for (int
k = 0; k < j; k++) s +=
Ai[k] * Aj[k]; Ai[j] =
div[j] * (Ai[j] - s); } float s = 0; for (int k =
0; k < i; k++) s +=
Ai[k] * Ai[k]; const float aii = (Ai[i] - s); Ai[i]
= (aii <=
sqrt(EPS)) ? (1/EPS) : sqrt(aii); div[i]
= (aii <= sqrt(EPS)) ? (0) : (1/sqrt(aii)); } |
However, the routine above is exceptionally slow
for large matrices. We rather have to use divide and
conquer
to recursively split the matrix into halves, then
use STRSM and SSYRK to perform a Schur Complement.
|
|
We first perform Cholesky on the top left corner.
Then, we solve with STRSM then use SSYRK.
|
|
We then have the transformed matrix:
|
|
We then perform Cholesky on the bottom right corner.
|
|
We can do this recursively. The main benefit is in
the old routine, Cholesky / STRSM / SSYRK takes P^3 time.
Well Cholesky 1/3P^3 FLOPS, STRSM and SSYRK both
1/2P^3 FLOPS.
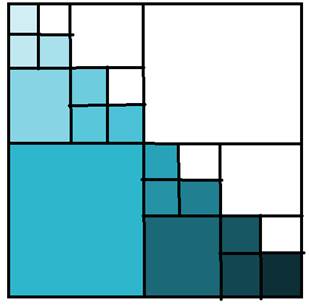
If we do a simple calculation of depth = 3, we will
find the following FLOPS:
|
|
This means the summation becomes:
|
|
However notice
how the original Cholesky takes 1/3n^3 FLOPS. So what’s
the big deal?
Parallelization! STRSM and SSYRK are extremely easy
to parallelize. In fact with K cores, assume a 1/K improvement.
Cholesky is much harder to parallelize,
since there are synchronization steps at each row. Assume a 1/log2(K)
improvement.
|
|
Now we see the benefit! An approx
sqrt(K) improvement. Ie if you had 16 cores, expect a
4 times improvement.
Jacobi
Preconditioning & Standardization
https://www.netlib.org
[SPSTRF]
https://www.gnu.org/software/gsl/doc/html/linalg.html#c.gsl_linalg_cholesky_scale
[Jacobi Preconditioning]
Also, to reduce the condition number of the matrix,
one can use Jacobi Preconditioning.
|
|
Another obvious way to counter large condition
numbers is to first standardize the matrix.
IF YOU DO
STANDARDIZATION TURN OFF JACOBI PRECONDITIONING!
|
|
However, we don’t want to
compute the standardized data since that’ll take a lot of memory.
We first precompute the reciprocal of the standard
deviations, then expand the above equation.
We also reset the reciprocal to 0 if the original standard
deviation was 0.
|
|
However
notice the following relation:
|
|
Which then we simplify the above to:
|
|
In other words, we do not need to create a new
memory space for the standardized X!
We can precompute XTX, then subtract the mean
factor, then multiply by the reciprocal standard deviation.
In all, the number of operations reduces from NP
FLOPS to now P^2 FLOPS.
Likewise
memory usage has reduced from an extra NP space to now virtually 0!!
|
|
Be careful of the N factor when you multiply
the 2 means!
However, sadly, for large dimensional data, ie p > n, the above method fails.
Since we have:
|
|
The above does NOT simplify sadly, so the only way
to compute the above is to actually compute
the standardized data. Though if memory is an issue,
we can compute column subsets then update XXT.
For matrix matrix
multiplication, we have a similar phenomenon:
|
|
So, we first copy and scale V by the scale factor.
Then perform a simple matrix vector multiply
To obtain the correction factor for each column.
This means we can reduce memory usage from NP to
now PK. This method is not
suggested if K > N.
|
|
However
in practice, especially for solving with Cholesky, one should NOT minus
the mean.
The removal of the mean can in fact REDUCE
accuracy!!
However
for PCA, mean removal is inevitable, or else clusters become incorrect.
So, for both XTX and XXT:
|
|
Ridge
Regression & LOOCV
http://pages.stat.wisc.edu/~wahba/ftp1/oldie/golub.heath.wahba.pdf [1979
Generalized Cross Validation for Choosing a Good Ridge Parameter]
https://arxiv.org/abs/1803.03635 [2018
The Lottery Ticket Hypothesis]
Ridge regression is particularly useful for reducing
the norm of the parameters.
It in fact has a closed form solution:
|
|
For underdetermined systems, Cholesky can become
rank deficient, and fail. So we have to add regularization.
A good heuristic:
|
|
This is mostly guaranteed to attain good accuracy.
In fact, this was the tolerance factor from LAPACKs pivoted Cholesky (SPSTRF).
We showcase how our choice of the optimum regularization
performs:
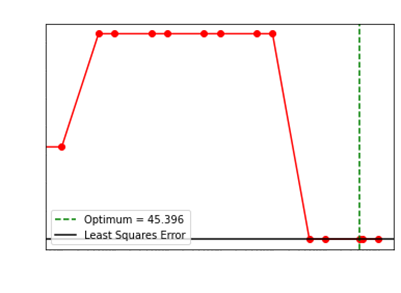
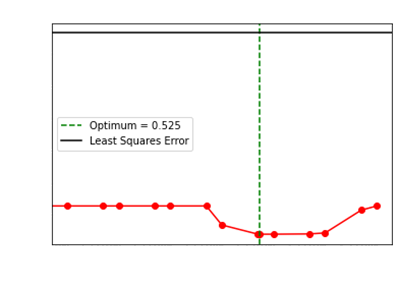

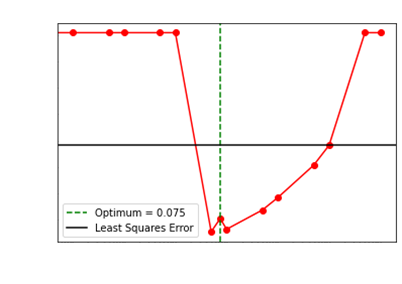

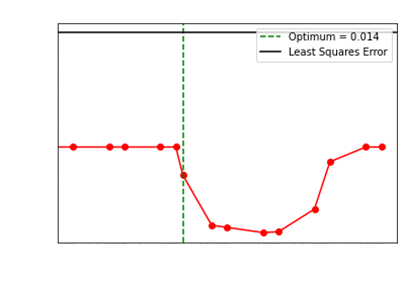
Levenberg
Marquardt Modified Correction
https://en.wikipedia.org/wiki/Levenberg%E2%80%93Marquardt_algorithm [Wikipedia]
Sadly for
very ill conditioned matrices, simply using stable Cholesky above still fails to
converge at all.
So we invoke Levenberg
Marquardt’s correction (ie the LM algorithm).
Instead of a simple Ridge regularization, we scale
according to the diagonal elements.
|
|
However
this method disregards collinear columns. If 2 columns are perfectly corrected,
LM will still fail.
So, we inject some deterministic jitter.
|
|
Now, we can set the regularization parameter to a small number ie epsilon * 10. Choosing epsilon
causes the addition to turn to not updating anything.
We then shift the brackets around.
|
|

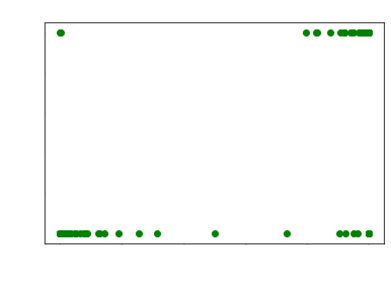
However
for extremely ill conditioned matrices, even the LM algorithm fails. So, we use
our optimal L2 regularization plus the LM
algorithm to make the final system always converge.
|
|
Copyright Daniel Han 2024. Check out Unsloth!